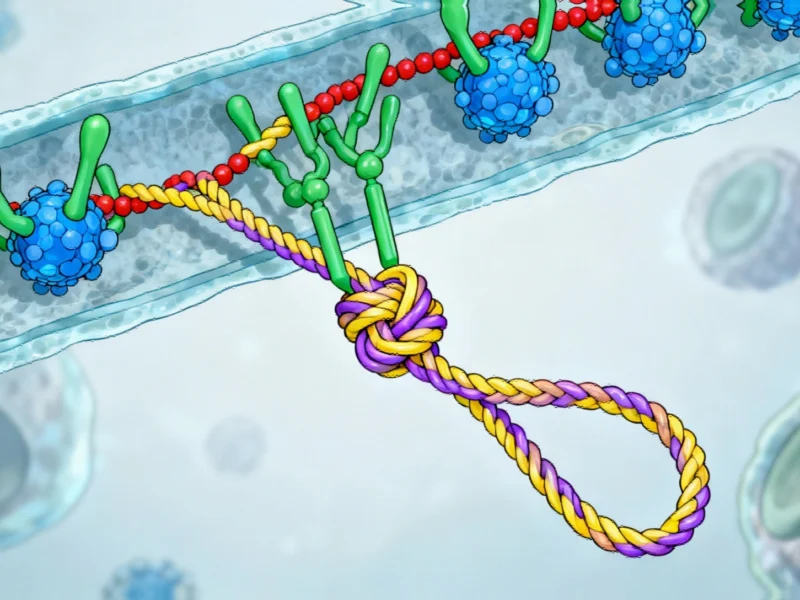
In the relentless pursuit of next-generation therapeutics for cancer and infectious diseases, researchers are turning to one of nature’s most structurally sophisticated molecules: lasso peptides. These remarkable compounds, characterized by their unique slip-knot configuration, offer unprecedented stability and diverse biological activities that make them ideal candidates for pharmaceutical development. A groundbreaking AI language model is now advancing lasso peptide therapeutic discovery through innovative computational approaches that could revolutionize how we engineer these complex molecules.
The recent development of LassoESM by researchers at the Carl R. Woese Institute for Genomic Biology represents a quantum leap in peptide engineering. Published in Nature Communications, this specialized large language model addresses the unique challenges posed by lasso peptides’ intricate structures, which have previously confounded conventional protein prediction algorithms like AlphaFold.
The Structural Marvel of Lasso Peptides
Lasso peptides are natural products synthesized by bacteria through a fascinating biosynthetic process. Ribosomes construct chains of amino acids that specialized biosynthetic enzymes then fold into distinctive slip-knot configurations. This natural engineering produces thousands of variants, many exhibiting potent antibacterial, antiviral, and anticancer properties.
“There are striking opportunities to use lasso peptides in drug discovery, from targeting receptors to developing stable oral therapeutics,” explained Doug Mitchell, Director of the Vanderbilt Institute for Chemical Biology and co-leader of the study. “By building a dedicated language model for these molecules, we’ve created a tool that helps us unlock these possibilities far more efficiently.”
Why Conventional AI Falls Short
While machine learning has become indispensable for pattern recognition in large biological datasets, existing protein prediction platforms struggle with lasso peptides’ unique architecture. “Because of the unique structure of the lasso peptide, none of the current AI programs actually work in terms of doing a structure prediction,” said project co-leader Diwakar Shukla, professor of chemical and biomolecular engineering at the University of Illinois Urbana-Champaign.
The challenge lies in the scarcity of experimentally labeled data and the complexity of enzyme-peptide substrate interactions. Standard protein language models, trained on conventional amino acid sequences and three-dimensional structures, lack the specificity required to accurately predict lasso peptide behavior.
The LassoESM Innovation
LassoESM represents a tailored solution to this computational gap. “We developed LassoESM, a lasso peptide-tailored protein language model, to capture peptide-specific features that are often missed by generic protein language models,” said Xuenan Mi, who recently earned her Ph.D. in Shukla’s research group.
The research team employed a multi-faceted approach, beginning with bioinformatics methods to identify thousands of lasso peptide sequences produced by various microorganisms. After manual validation to ensure data quality, the researchers applied masked language modeling techniques. “We learned the language of those lasso peptides using masked language modeling, which is where you hide part of the peptide, and then you try to predict the other half,” Shukla explained.
Practical Applications and Clinical Potential
The integration of computational expertise from Shukla’s group with experimental data from Mitchell’s laboratory enabled LassoESM to perform numerous practical prediction tasks. A key application involves identifying compatible lasso peptide and lasso cyclase pairs—the enzymes responsible for the crucial knot-forming step in biosynthesis.
“We built the models to predict which lasso cyclase could actually form a lasso peptide using only the sequence of amino acids in a peptide,” Shukla said. “If we can understand the substrate scope or we can engineer lasso cyclases, then we can potentially make any peptide into a lasso.” This capability represents a significant advancement, as these enzyme-substrate interactions have traditionally been difficult to predict.
The implications extend beyond basic research, potentially influencing broader technological landscapes. Just as new economic frameworks are emerging to address global manufacturing challenges, computational tools like LassoESM are creating new paradigms for pharmaceutical development.
Computational Synergies and Future Directions
The success of LassoESM highlights how specialized computational approaches are transforming multiple scientific domains. Similar to how AMD’s Zen 5 architecture addresses critical computational challenges in processor design, tailored language models are solving specific problems in biochemical engineering.
The research team demonstrated that LassoESM enables accurate prediction of various lasso peptide properties even with limited training data. “This work provides a powerful AI-driven tool to accelerate the rational design of functional lasso peptides for biomedical and industrial applications,” Mi confirmed.
Future developments will expand the model’s capabilities to include tailor-made language models for other peptide natural products and engineered lasso peptides targeting specific proteins. The computational infrastructure supporting this research reflects broader trends in high-performance computing, reminiscent of advances seen in industrial computing systems handling complex processing requirements.
Interdisciplinary Collaboration and Technological Infrastructure
The project’s success underscores the importance of interdisciplinary collaboration and robust computational resources. “Thanks to access to powerful computing resources on our campus and interdisciplinary collaboration opportunities provided by the MMG theme at Carl R. Woese Institute for Genomic Biology,” Shukla noted, acknowledging the contributions of team members and collaborators.
This research approach mirrors innovations in other fields where specialized computational tools are driving discovery. The development of autonomous systems, similar to next-generation AUV deployment advancing polar research, demonstrates how targeted computational solutions are enabling breakthroughs across scientific disciplines.
The methodology also shares conceptual parallels with materials science innovations, such as polymers that spontaneously develop chirality through sophisticated molecular interactions, and breakthrough fluorescent molecules defying conventional design principles.
Transforming Therapeutic Discovery
LassoESM represents more than just another computational tool—it signifies a fundamental shift in how we approach complex biological structures. By learning the intricate “language” of lasso peptides, researchers can now accelerate the design and optimization of these promising therapeutic candidates.
As the team continues to refine and expand their model, the potential applications grow increasingly exciting. The ability to engineer lasso peptides with specific targeting capabilities could open new frontiers in precision medicine, while the underlying computational framework could be adapted to other challenging molecular structures.
This research demonstrates how the convergence of computational power, specialized algorithms, and biological insight is creating new possibilities in drug discovery—possibilities that were previously constrained by the limitations of conventional prediction methods.
Based on reporting by {‘uri’: ‘phys.org’, ‘dataType’: ‘news’, ‘title’: ‘Phys.org’, ‘description’: ‘Phys.org internet news portal provides the latest news on science including: Physics, Space Science, Earth Science, Health and Medicine’, ‘location’: {‘type’: ‘place’, ‘geoNamesId’: ‘3042237’, ‘label’: {‘eng’: ‘Douglas, Isle of Man’}, ‘population’: 26218, ‘lat’: 54.15, ‘long’: -4.48333, ‘country’: {‘type’: ‘country’, ‘geoNamesId’: ‘3042225’, ‘label’: {‘eng’: ‘Isle of Man’}, ‘population’: 75049, ‘lat’: 54.25, ‘long’: -4.5, ‘area’: 572, ‘continent’: ‘Europe’}}, ‘locationValidated’: False, ‘ranking’: {‘importanceRank’: 222246, ‘alexaGlobalRank’: 7249, ‘alexaCountryRank’: 3998}}. This article aggregates information from publicly available sources. All trademarks and copyrights belong to their respective owners.